Introduction
Sickle cell disease (SCD) is a chronic condition with significant morbidity in children and early adult mortality. Identifying high-risk SCD populations has been challenging due to limited population cohorts of children and adults. Understanding the clinical course of SCD in both children and adults is crucial for informed treatment decisions around disease-modifying therapy vs. curative and transformative therapies. Recent use of Medicaid, Medicare, and private insurer claims datasets has helped define high-risk populations and estimate mortality, but they have limitations in clinical record depth. In contrast, electronic health record (EHR)-based datasets offer a better option for assessing the impact of disease-modifying therapy and allow comprehensive mortality risk prediction analyses, using high-dimensional laboratory data.
Objective
Previously, using the data warehouse at a single institution (Vanderbilt University School of Medicine) with data collected over 18 years, members of our team developed a contemporaneous cohort of children and adults with SCD. To do this our team developed a dual-approach algorithm that combined patients categorically selected by diagnosis using ICD-10 (International Classification of Diseases -10) codes and by laboratory data including hemoglobin fractionation values. The team estimated the median survival in adults with SCD and predicted mortality risk based on disease-modifying therapy and insurance status. We expanded this work by testing the hypothesis that we can first apply the laboratory portion of the Vanderbilt algorithm to identify children and adults with SCD living in the United States from the national Cerner Learning Health Network (Cerner LHN) EHR dataset.
Methods
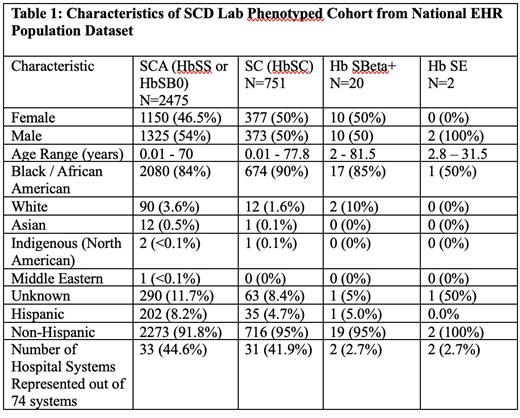
The Cerner LHN is a nationwide dataset comprised of 88 million patients from over 74 hospital systems across 37 states (as of September 17, 2021). We used data from the Cerner LHN collected between 8/26/2002 and 12/13/2022 for our analysis. We extracted laboratory data elements required for the Vanderbilt algorithm including hemoglobin fractionation data with HbS values as well as total hemoglobin timepoints and blood transfusion time frames. The results of the laboratory portion of the Vanderbilt algorithm predict genotype of sickle cell disease in the following categories: SCA (HbSS or HbSB 0 ), SC (Hb SC), HbSBeta + and HbSE. There were 5,827 patients identified with these laboratory components necessary for the algorithm. We applied an updated python script that detailed the logic from the Vanderbilt algorithm on the 5,827 patients and we identified 3,248 children and adults with SCD. Patients were then evaluated for demographics and hospital systems coverage within the Cerner LHN.
Results
Black/African Americans made up 2947 patients (90.7%) of the SCD cohort. There were 1325 (54%) male and 1150 (46.5%) female patients with SCA within the phenotyped cohort. Patients with SCA were collected from 44.6% of the Cerner LHN health systems.
Conclusions
The population was 90.7% Black/African American as expected in patients carrying a diagnosis of SCD. This contemporaneous national cohort represents 44.6% of the health systems within the Cerner LHN which denoted wide-reaching patient sampling. These findings show that the laboratory portion of the Vanderbilt SCD algorithm can, in an automated fashion, establish a phenotyped SCD cohort of children and adults in a national EHR system. We plan to power the total number by adding the ICD-10 portion of the algorithm and estimating mortality in the final cohort. Further, we will validate combined algorithm by chart review.
Disclosures
DeBaun:FORMA: Consultancy; Novartis Pharmaceuticals Corporation: Other: Study steering committee member; Global Blood Therapeutics: Membership on an entity's Board of Directors or advisory committees, Research Funding; Vertex/CRISPR: Membership on an entity's Board of Directors or advisory committees.